Broadly, I am interested in understanding how somatic mutation and epigenetic regulation of genomes contributes to cellular differentiation, speciation, and disease. My exploration of the [epi]genome has often involved implementation of custom algorithms and pipelines for analysis of next-generation sequencing technologies, and occasionally the development of new statistical or computational theory. Since graduating from USC, I have focused primarily on applying my background to understanding the intricacies of and evaluating potential therapies for a broad range of diseases.
 |
|---|
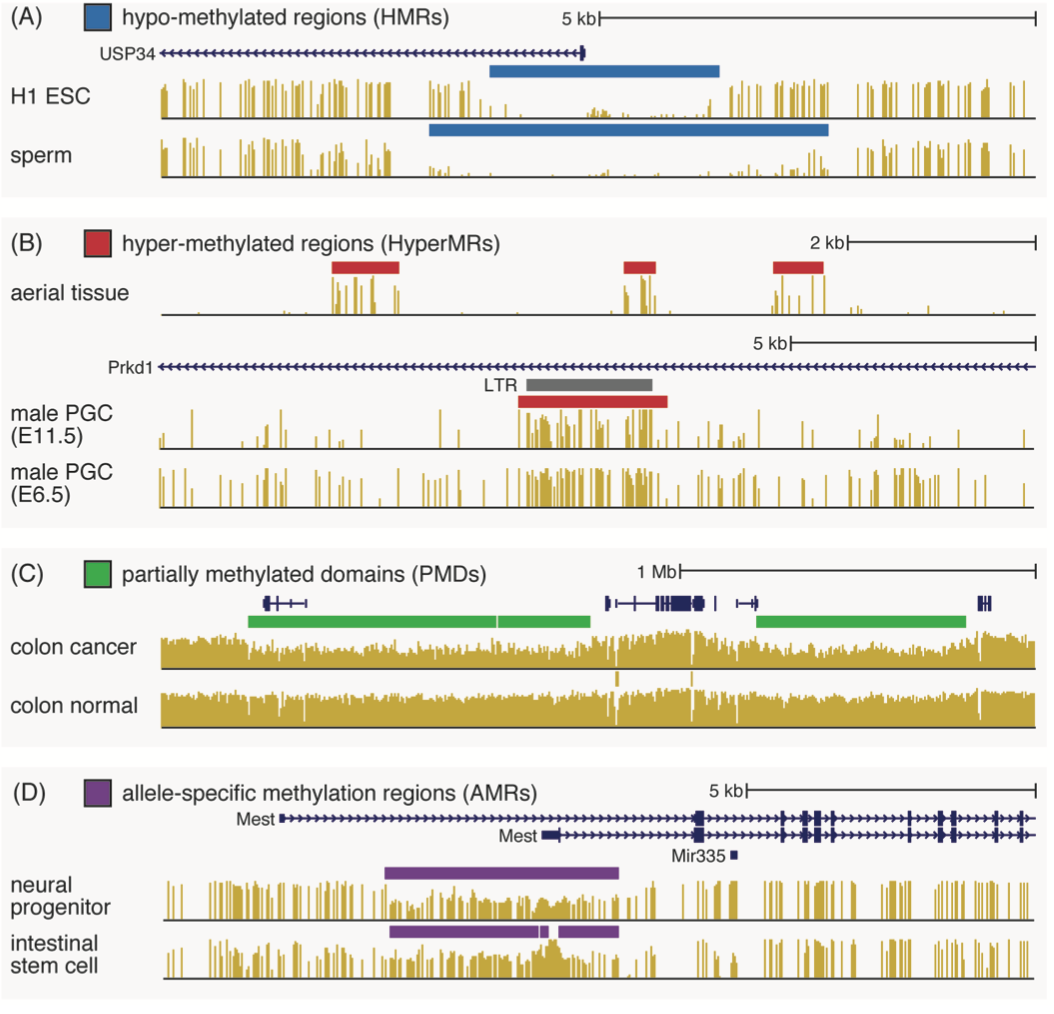
| Major landmarks of the methylome. (A) HMRs, kilobase-scale regions of near-complete hypomethylation that frequently co-locate with transcription start sites, and whose absence correlates with transcriptional silencing. (B) HyperMRs, which serve roughly the opposite function of HMRs and exist in plant methylomes. (C) Partially methylated domains: megabase scale regions of lower-than-average methylation that form in tissues with long mitotic history like cancer. These were a major part of my Ph.D. research. (D) AMRs, which mark regions of allele-specific methylation, typically at imprinting control regions and indicative of parent conflict in developmental cell types. |